|
bookmark this page - make qweas your homepage | |
| Help Center - What's New - Newsletter - Press | ||
| Get Buttons - Link to Us - Feedback - Contact Us |
| Home | Download | Store | New Releases | Most Popular | Editor Picks | Special Prices | Rate | News | FAQ |
|
|
Classifion 1.2 - User Guide and FAQScreenshots - More Details
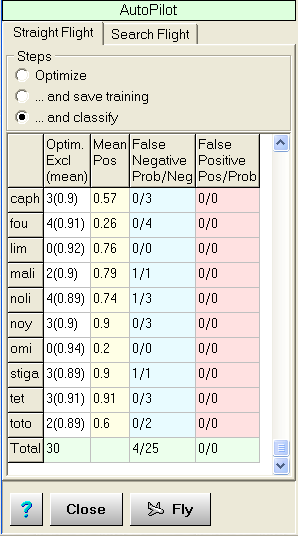
Where does it sit... Almost every mass-spectrometry lab is equipped with spectral-library search program with tens of thousands of spectra of pure compounds. The problems begin when you have very complex substances or using mass-spec methods different from those used in the spec-libraries. In some cases, you don't need to know what exactly is in the sample, but simply does it belong to a class of samples you already measured. In all these cases, you need to use classification software. But most of them are general purposed and require certain knowledge of the statistical methods used. Here Classifion sits, offering you a classification with specialized for mass-spectrometry environment and automation allowing you to "avoid the depths of contemporary statistics", so you can concentrate on your immediate work. How does it work... Classifion needs to be "trained" using several measured spectra per substance. The "training" itself could be conducted automatically or manually (supervised). The aim of the training is to extract substance specific information based on statistical characteristics, but not on the interpretation of the mass-spectra. Having some knowledge on the mass-spectra structure could improve classification precision. Mathematically speaking the extraction is down to reducing the dimensionality of the spectra variable space (to each mass corresponds one dimension). PCA is well-known such as technique, which offers other advantages as ordering the principal component by "significance" (useful for noise reduction). Classifion can train itself entirely on autopilot. Just import your data, run Autopilot, sit back and enjoy the view. The results will show you how successful the training was, so in case of problem you should re-examine your data or do supervised training. The feature which makes Classifion better than the most of PCA software out there, is the use of proper factor space for each substance separately, instead of traditional approach of using common factor space for all substances. That improves the classification precision and makes the work with Classifion similar to the spectral-library search software. The software... Classifion is designed to work as stand-alone application as well as out-of-process COM-server. The latter allows Classifion to be used remotely from another application combining or not the remote access with user access. Step-by-step example The objective: based on their mass-spectra to classify an unknown sample to be (or not) one of number of known substances. That example follows the shortest, most automated way for classification. That will work in the majority of the cases with decent data. Classifion will estimate the quality of your data for you.
Screenshots - More Details |
|
Search -
Download -
Store -
Directory -
Service -
Developer Center
© 2006 Qweas Home - Privacy Policy - Terms of Use - Site Map - About Qweas |


 Download Site 1
Download Site 1 Buy Now $145.00
Buy Now $145.00